
Z - 1
A computational platform for data support, integration and management
Dr. David H. Meyer

Thorsten Hoppe
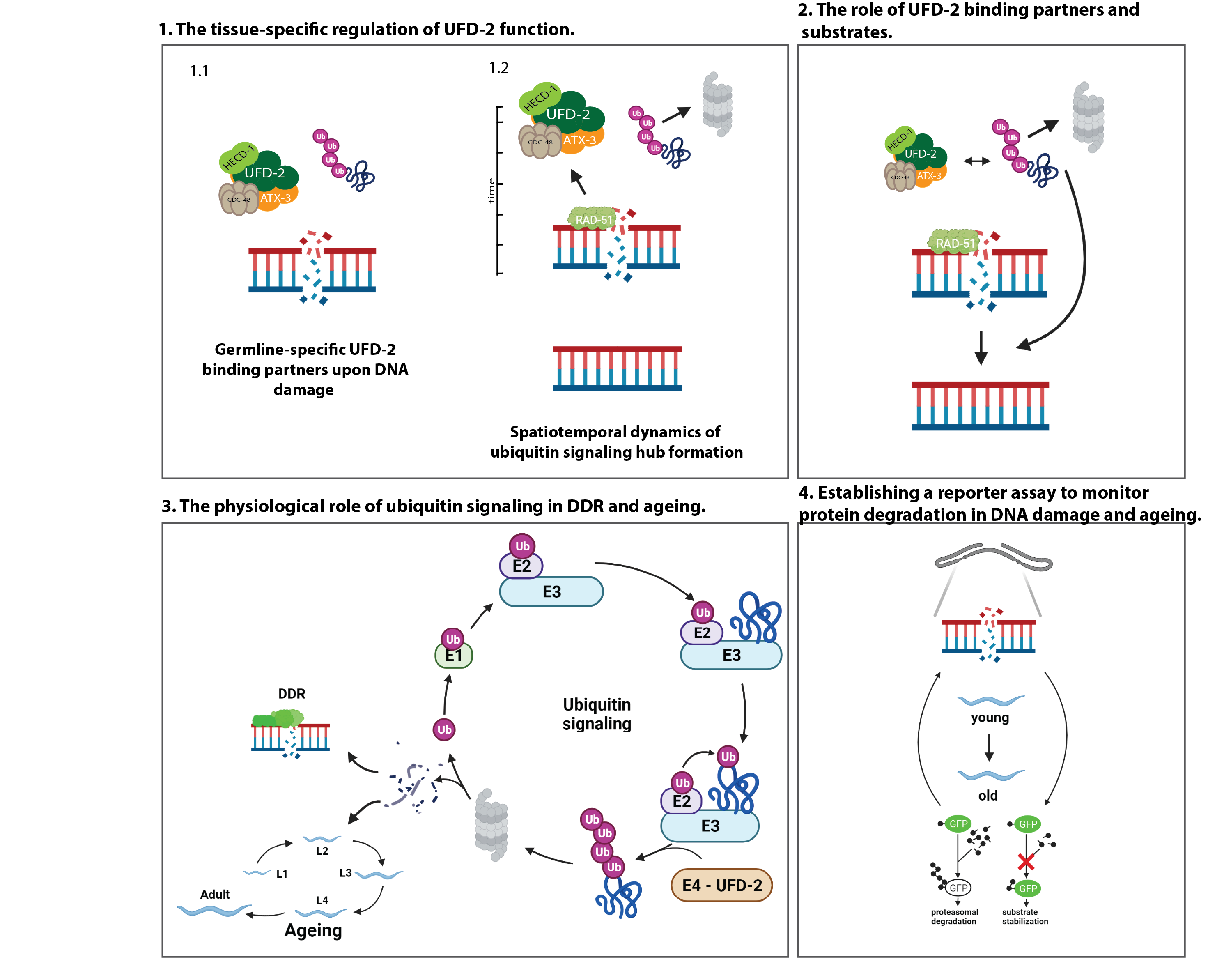
Repair of DNA double-strand breaks (DSBs) is tightly regulated by ubiquitin modification. Ubiquitin-dependent proteolysis is known to decrease with age, correlating with increased aggregation of damaged proteins. In contrast, the physiological role of the ubiquitin/proteasome-system (UPS) in age-related DNA damage accumulation and genome instability remains largely unclear. Our recent work has uncovered a central role of the conserved ubiquitin ligase UFD-2 in DSB repair, which is important for germline function, fecundity, and lifespan in C. elegans. UFD-2 is a specialized enzyme that triggers proteolysis by converting a nondegradable ubiquitin signal into a signal destined for substrate turnover by the 26S proteasome. Following DNA damage, UFD-2 forms focal accumulations in the germline that persist until homologous recombination is complete, reflecting the intricate coordination between ubiquitin-dependent regulation and DSB repair. The UFD-2 foci also contain the ubiquitin-selective segregase CDC-48/p97 and the deubiquitylation enzyme ATX-3/Ataxin-3, both of which have conserved functions in chromatin-associated protein degradation, DSB repair, and longevity. The main goal of the proposed research is to understand the physiological role of ubiquitin signaling in genome stability during development and aging. The project will address and systematically analyze the spatiotemporal coordination of ubiquitin-dependent DSB repair: the tissue-specific regulation of UFD-2 function and the physiological role of ubiquitin signaling and DNA damage response (DDR) in aging. To this end, in vitro and in vivo ubiquitylation assays, biotinylation-based protein-protein interaction mapping will be performed in combination with large-scale proteomics, CRISPR/Cas9 gene editing, time-lapse microscopy, and lifespan measurements will be performed.
PROJECT RELATED PUBLICATIONS
Articles published by outlets with scientific quality assurance, book publications, and works accepted for publication but not yet published. (*co-corresponding author)

