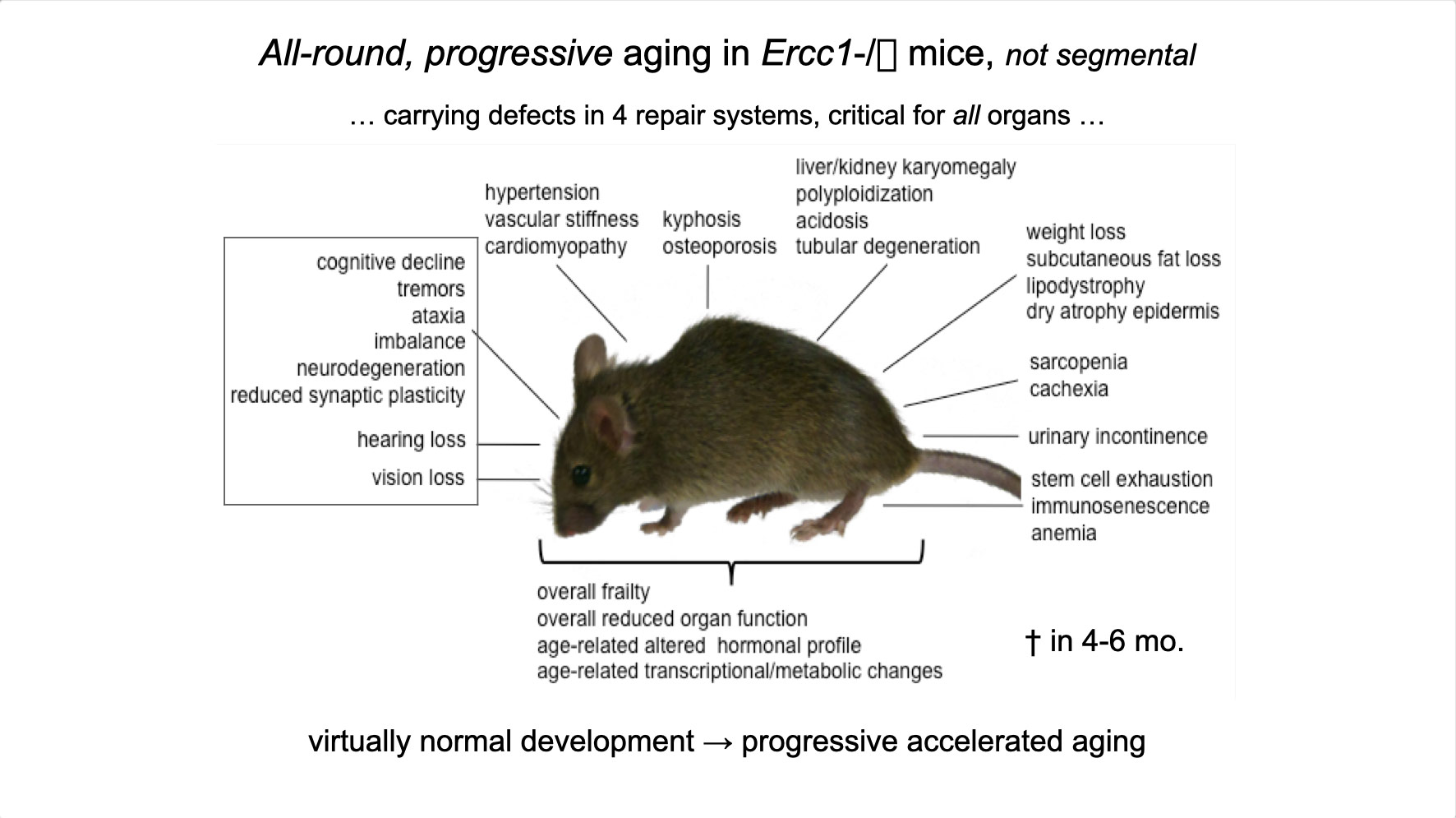
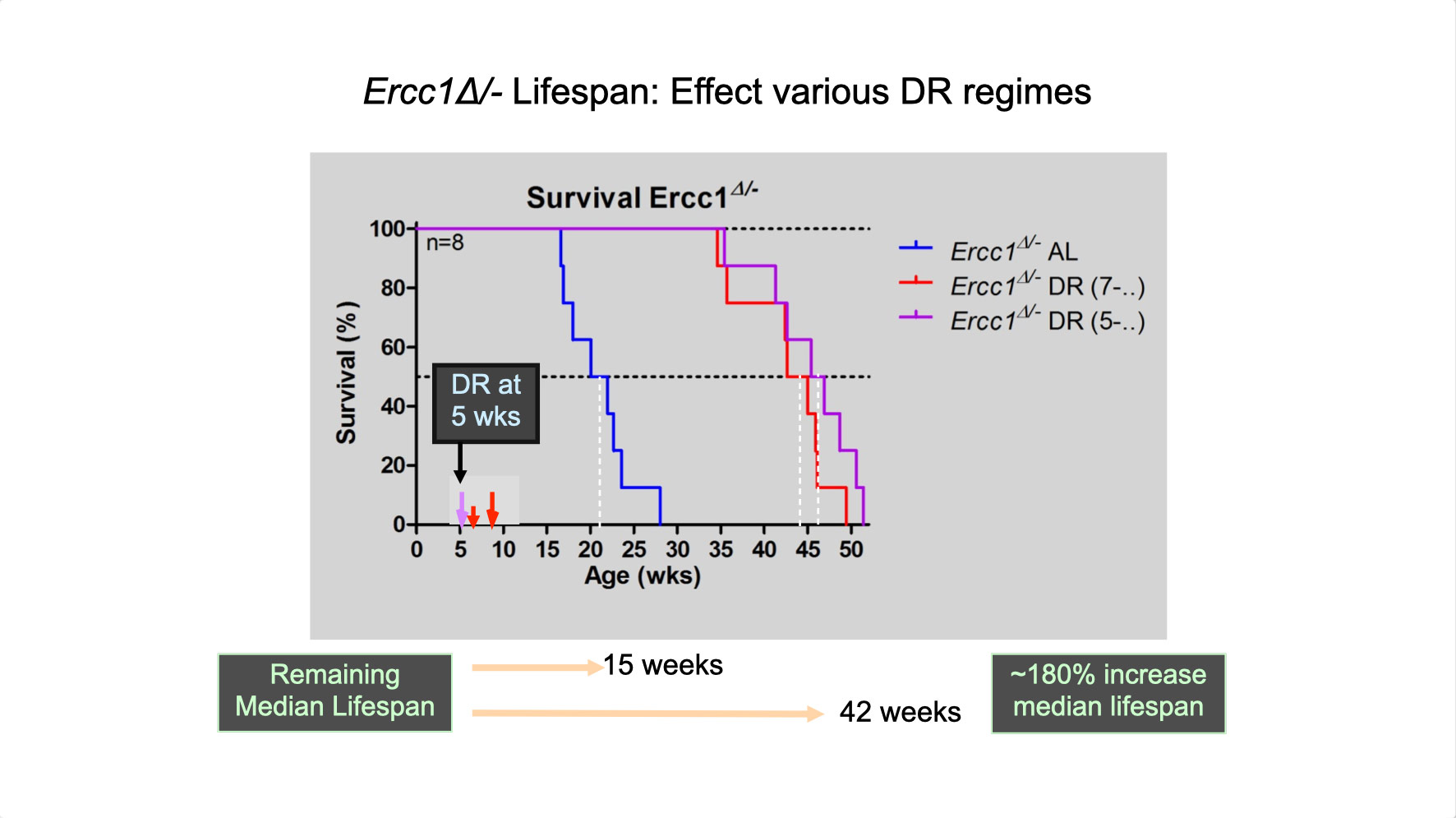
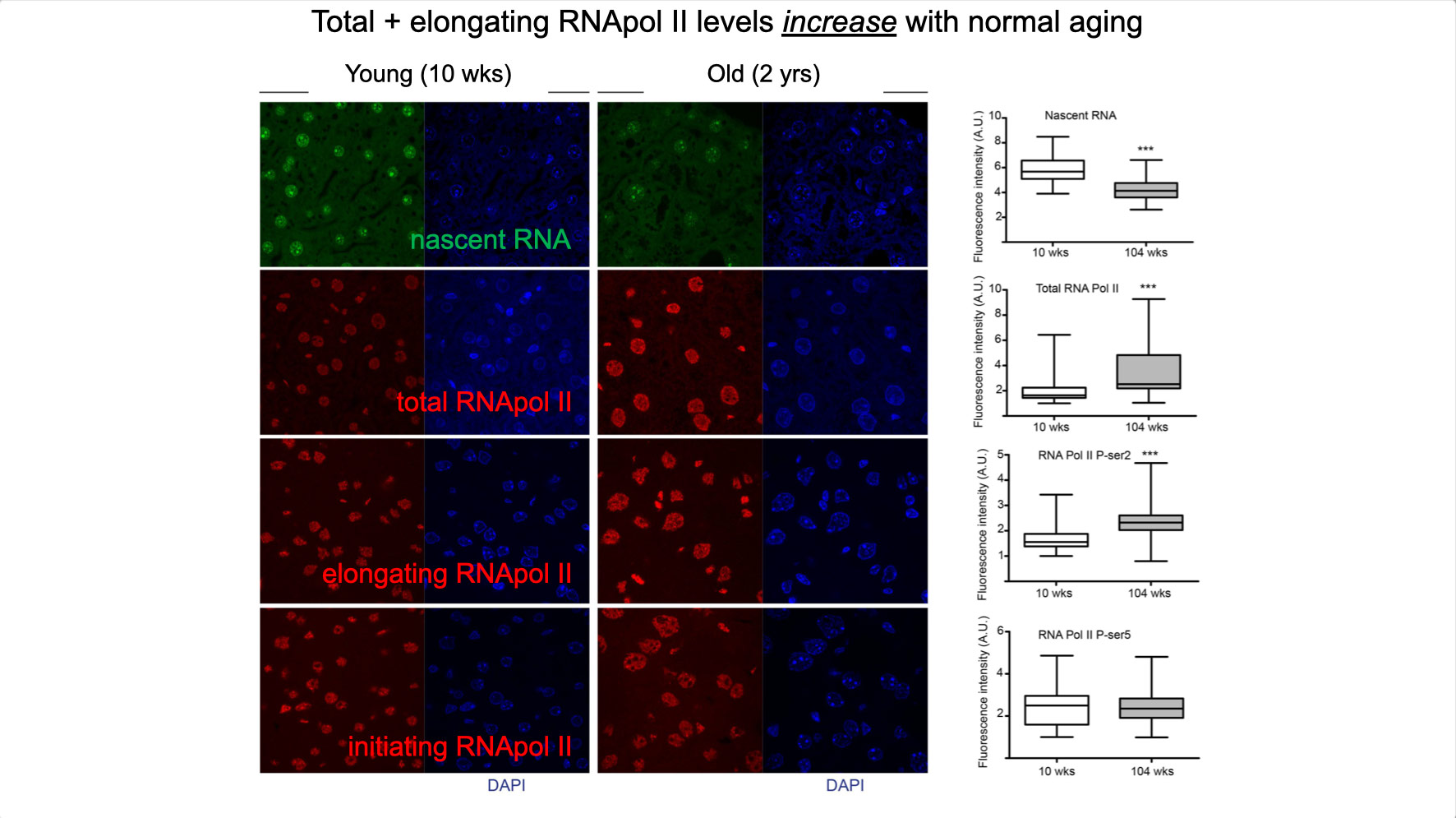
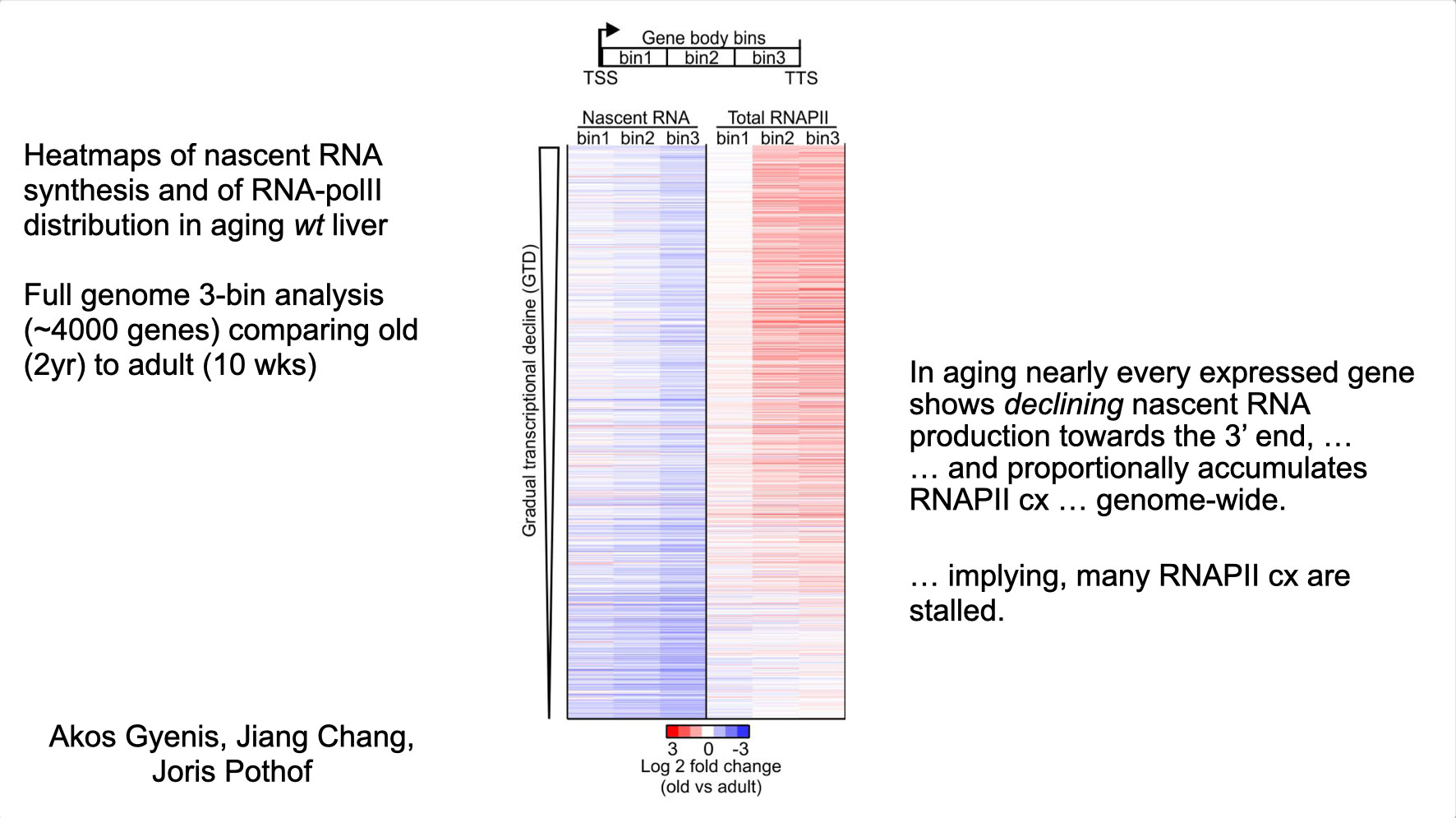
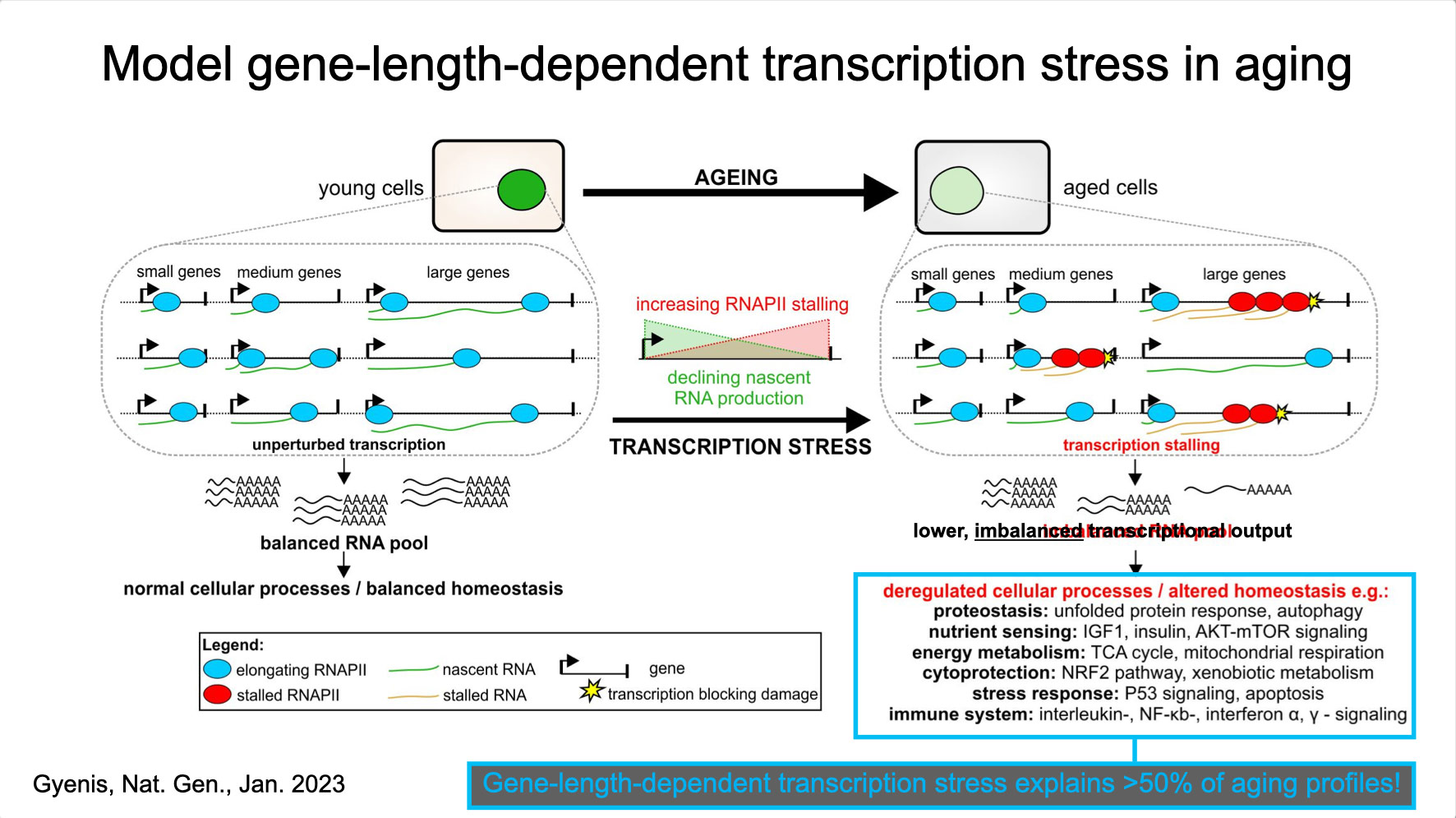
To properly maintain cell functioning and homeostasis and respond to different environmental stimuli, cells require intact genome and proficient chromatin maintenance. Gene expression profiling has identified numerous processes, which are altered in aging, but how these changes arise is largely unknown. We combined nascent RNA sequencing and RNA polymerase II ChIP-seq to explore underlying mechanisms influencing gene expression changes in wildtype aged mice. We found that in 2-year-old livers, 40% of elongating RNApolII complexes are stalled, drastically lowering productive transcription at a rate of 0,35%/kb and skewing transcriptional output in a gene-length-dependent fashion. This ‘transcription stress’ (TS) is directly caused by endogenous DNA damage and explains the majority of gene expression changes in aging in many organs, affecting aging hallmark pathways such as nutrient sensing, autophagy, proteostasis, energy metabolism, immune function and cellular stress resilience and is evolutionary conserved from nematodes to man. These findings disclose how DNA damage functionally underlies major aspects of normal aging. Although aging is universal, not every tissue and cell type ages the same rate. Understanding the key mechanisms responsible for tissue-specific aging could help finding effective anti-aging strategies. Currently, dietary restriction (DR), is the most robust intervention delaying age-associated pathologies in numerous tissues and species, but how it works is still largely unknown. Here, we propose a comprehensive multidisciplinary (in vivo, in vitro and in silico) approach to investigate how age-related accumulating DNA damage influences homeostasis in 4 key organs (heart, muscle, brain and kidney) and how DR counteracts aging. We aim to 1. characterizing the genomic, transcriptional and metabolic burden of increasing DNA damage and TS in the above postmitotic tissues in progeroid repair mutants and wt mice. 2. Examine the effect of DR-induced metabolic redesign on TS, DNA damage and organismal health. 3. Identify key mechanisms and signaling pathways responsible for DR-based metabolic redesign.
PROJECT RELATED PUBLICATIONS
- Lans H, Hoeijmakers JHJ, Vermeulen W, Marteijn JA. The DNA damage response to transcription stress. Nat Rev Mol Cell Biol. 2019 Sep 26. doi: 10.1038/s41580-019-0169-4. PMID: 31558824.
- Milanese C, Bombardieri CR, Sepe S, Barnhoorn S, Payán-Goméz C, Caruso D, Audano M, Pedretti S, Vermeij WP, Brandt RMC, Gyenis A, Wamelink MM, de Wit AS, Janssens RC, Leen R, van Kuilenburg ABP, Mitro N, Hoeijmakers JHJ, Mastroberardino PG. DNA damage and transcription stress cause ATP-mediated redesign of metabolism and potentiation of anti-oxidant buffering. Nat Commun. 2019 Oct 25;10(1):4887. doi: 10.1038/s41467-019-12640-5. PMID: 31653834
- B. Schumacher, J. Pothof, J. Vijg, J.H.J. Hoeijmakers. The central role of DNA damage in the ageing process. Nature. 2021 Apr;592(7856):695-703. doi: 10.1038/s41586-021-03307-7. Epub 2021 Apr 28. PMID: 33911272. (This review certifies our primary claim, initially met with strong resistance from the aging research field, that DNA damage is at the basis of systemic aging).
- Vougioukalaki M, Demmers J, Vermeij WP, Baar M, Bruens S, Magaraki A, Kuijk E, Jager M, Merzouk S, Brandt RMC, Kouwenberg J, Van Boxtel R, Cuppen E, Pothof J, Hoeijmakers JHJ. Different responses to DNA damage determine ageing differences between organs. Aging Cell. 2022 Apr;21(4):e13562. doi: 10.1111/acel.13562. Epub 2022 Mar 4. PMID: 35246937
- Gyenis A, Chang J, Demmers JJPG, Bruens ST, Barnhoorn S, Brandt R, Baar MP, Raseta M, Derks KWJ, Hoeijmakers JHJ, Pothof J. Genome-wide transcription stalling by DNA damage shapes the transcriptome in aging. Nature Genetics 2023 Feb;55(2):268-279, doi.org/10.1038/s41588-022-01279-6 2023. Epub 2023 Jan 19. (Discovery of DNA damage-driven qualitative and quantitative decline of genome-wide transcription underlying most hallmarks of aging).
- Martine de Boer; Maaike te Lintel Hekkert; Jiang Chang; Bibi S. van Thiel; Leonie Martens; Maxime M. Bos; Marion G.J. de Kleijnen; Yanto Ridwan; Yanti Octavia; Elza D. van Deel; Lau A. Blonden; Renata M.C. Brandt; Sander Barnhoorn; Paula K. Bautista-Niño; Ilona Krabbendam-Peters; Rianne Wolswinkel; Banafsheh Arshi; Mohsen Ghanbari; Christian Kupatt; Leon J. de Windt; A. H. Jan Danser; Ingrid van der Pluijm; Carol Ann Remme; Monika Stoll; Joris Pothof; Anton J.M. Roks; Maryam Kavousi; Jeroen Essers; Jolanda van der Velden; Jan H.J. Hoeijmakers; Dirk J. Duncker. DNA repair in cardiomyocytes is critical for maintaining cardiac function in mice. Aging Cell. 2023;00:e13768. https://doi.org/10.1111/acel.13768 PMID: 36756698